This function takes a series of MS/MS spectra, selects the most intense scan and extracts the MS/MS spectra from it. Additionally, it plots the MS/MS EIC chromatogram and colors the most intense scan with red circle, and the precursor ion with a blue diamond
Arguments
- spec
a data frame with the MS/MS spectra
- verbose
a boolean indicating if the MS/MS EIC chromatogram is displayed
- out_list
a boolean expressing if the output is a list containing the MS/MS spectra plus the EIC chromatogram (verbose = TRUE), or only the data frame with the MS/MS spectra (verbose = FALSE).
Examples
# Importing the Spectrum of Procyanidin A2 in negative ionzation mode
# and 20 eV as the collision energy
ProcA2_file <- system.file("extdata",
"ProcyanidinA2_neg_20eV.mzXML",
package = "MS2extract"
)
# Region of interest table (rt in seconds)
ProcA2_data <- data.frame(
Formula = "C30H24O12", Ionization_mode = "Negative",
min_rt = 163, max_rt = 180
)
# Importing MS/MS data
ProcA2_raw <- import_mzxml(ProcA2_file, ProcA2_data)
#> • Processing: ProcyanidinA2_neg_20eV.mzXML
#> • Found 1 CE value: 20
#> • Remember to match CE velues in spec_metadata when exporting your library
#> • m/z range given 10 ppm: 575.11376 and 575.12526
# Extracting most intense scan ----
# Returning plot + MS2 spectra
extract_MS2(ProcA2_raw)
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
#> Warning: `position_stack()` requires non-overlapping x intervals.
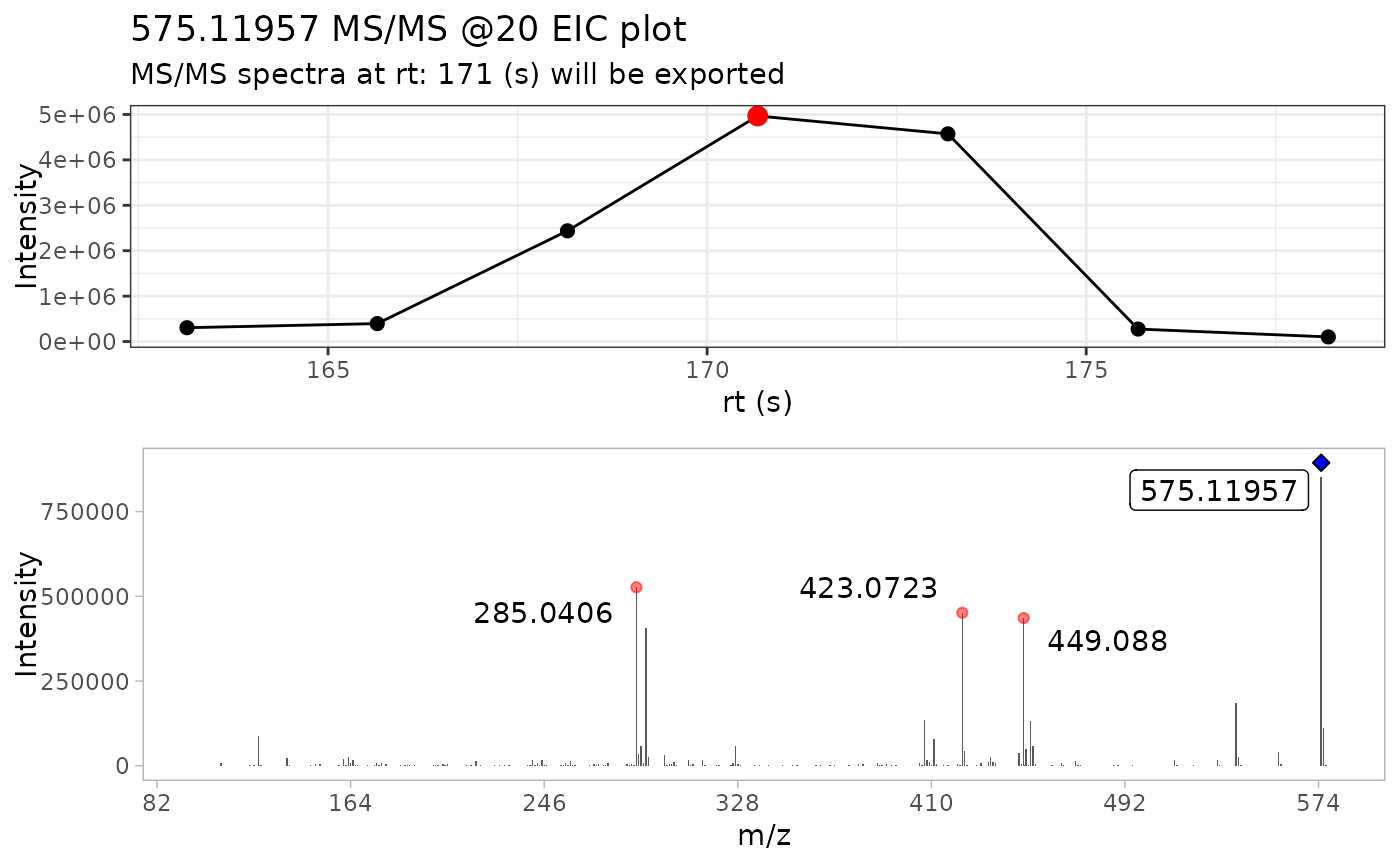 #> # A tibble: 4,561 × 6
#> # Groups: Formula, CE [1]
#> mz intensity mz_precursor rt CE Formula
#> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 100. 40.1 575. 171. 20 C30H24O12
#> 2 100. 23.3 575. 171. 20 C30H24O12
#> 3 100. 15.8 575. 171. 20 C30H24O12
#> 4 101. 255. 575. 171. 20 C30H24O12
#> 5 101. 11 575. 171. 20 C30H24O12
#> 6 102. 91.5 575. 171. 20 C30H24O12
#> 7 103. 9 575. 171. 20 C30H24O12
#> 8 104. 21.2 575. 171. 20 C30H24O12
#> 9 104. 91.1 575. 171. 20 C30H24O12
#> 10 104. 14.7 575. 171. 20 C30H24O12
#> # ℹ 4,551 more rows
# Returning MS/MS spectra only
extract_MS2(ProcA2_raw, out_list = FALSE)
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
#> Warning: `position_stack()` requires non-overlapping x intervals.
#> # A tibble: 4,561 × 6
#> # Groups: Formula, CE [1]
#> mz intensity mz_precursor rt CE Formula
#> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 100. 40.1 575. 171. 20 C30H24O12
#> 2 100. 23.3 575. 171. 20 C30H24O12
#> 3 100. 15.8 575. 171. 20 C30H24O12
#> 4 101. 255. 575. 171. 20 C30H24O12
#> 5 101. 11 575. 171. 20 C30H24O12
#> 6 102. 91.5 575. 171. 20 C30H24O12
#> 7 103. 9 575. 171. 20 C30H24O12
#> 8 104. 21.2 575. 171. 20 C30H24O12
#> 9 104. 91.1 575. 171. 20 C30H24O12
#> 10 104. 14.7 575. 171. 20 C30H24O12
#> # ℹ 4,551 more rows
# Returning MS/MS spectra only
extract_MS2(ProcA2_raw, out_list = FALSE)
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
#> Warning: `position_stack()` requires non-overlapping x intervals.
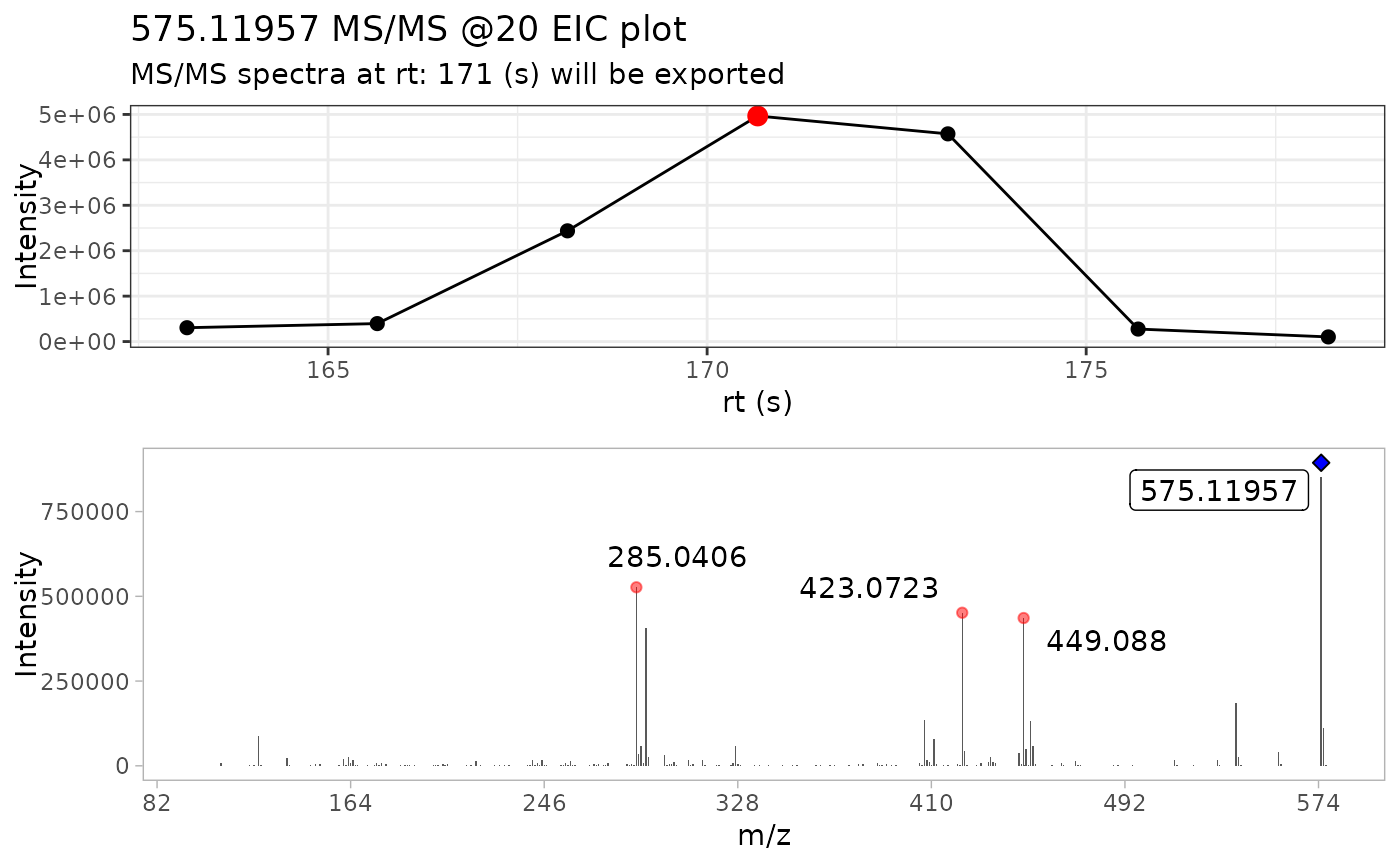 #> # A tibble: 4,561 × 6
#> # Groups: Formula, CE [1]
#> mz intensity mz_precursor rt CE Formula
#> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 100. 40.1 575. 171. 20 C30H24O12
#> 2 100. 23.3 575. 171. 20 C30H24O12
#> 3 100. 15.8 575. 171. 20 C30H24O12
#> 4 101. 255. 575. 171. 20 C30H24O12
#> 5 101. 11 575. 171. 20 C30H24O12
#> 6 102. 91.5 575. 171. 20 C30H24O12
#> 7 103. 9 575. 171. 20 C30H24O12
#> 8 104. 21.2 575. 171. 20 C30H24O12
#> 9 104. 91.1 575. 171. 20 C30H24O12
#> 10 104. 14.7 575. 171. 20 C30H24O12
#> # ℹ 4,551 more rows
#> # A tibble: 4,561 × 6
#> # Groups: Formula, CE [1]
#> mz intensity mz_precursor rt CE Formula
#> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 100. 40.1 575. 171. 20 C30H24O12
#> 2 100. 23.3 575. 171. 20 C30H24O12
#> 3 100. 15.8 575. 171. 20 C30H24O12
#> 4 101. 255. 575. 171. 20 C30H24O12
#> 5 101. 11 575. 171. 20 C30H24O12
#> 6 102. 91.5 575. 171. 20 C30H24O12
#> 7 103. 9 575. 171. 20 C30H24O12
#> 8 104. 21.2 575. 171. 20 C30H24O12
#> 9 104. 91.1 575. 171. 20 C30H24O12
#> 10 104. 14.7 575. 171. 20 C30H24O12
#> # ℹ 4,551 more rows