2 - Content description of the Phenolics MS/MS repository
Cristian Quiroz-Moreno, Jessica Coopestone
Horticulture and Crop Science Department, Ohio State University, USquirozmoreno.1@osu.edu
25 August 2025
Source:vignettes/Content.Rmd
Content.RmdPackage content description
This package is intended to archive the raw .mzMl files
of authentic analytical standards, as well as structured data frames
aimed to be used jointly with MS2extract, to
automate the process of creating in-house MS/MS libraries.
This database contains majorly phenolic compounds, aimed to decipher
and annotate the Rosacea metabolome. However, as phenolics are
expressed almost ubiquitously in plants, this database can be used in
multiples scenarios where researchers rationalize
PhenolicsDB could help to annotate metabolites present in
their dataset.
We grouped the total database in two batches. The batches are based on the instrument we used to acquire the MS/MS data. For the first batch, we used an Agilent QTOF 6545, and for the second batch, we used an Agilent QTOF 6546. In both batches, we collected targeted MS/MS using the chromatographic method reported in Bilbrey at al. (2022). We collected the data in negative and positive polarity, and we used 20 and 40 eV (CE), using collision induced dissociation (CID). Additionally, in the second batch, we also collected data at 60 and 80 eV (CE).
Again, this data package is intended to work with MS2extract, as
its structure meets the requirements in MS2extract,
where the read_dt tables enable importing the
.mzML files, while the met_metadata has
crucial information that is included in the final MS/MS library.
First Batch (Agilent QTOF 6545)
| Polarity | Collision Energy (eV) | read_dt table | met_metadata table |
|---|---|---|---|
| Positive | 20 | data("read_pos20_6545") |
data("metdt_pos20_6545") |
| Positive | 40 | data("read_pos40_6545") |
data("metdt_pos40_6545") |
| Negative | 20 | data("read_neg20_6545") |
data("metdt_neg20_6545") |
| Negative | 40 | data("read_neg40_6545") |
data("metdt_neg40_6545") |
Second Batch (Agilent QTOF 6546)
| Polarity | Collision Energy (eV) | read_dt table | metadata table |
|---|---|---|---|
| Positive | 20 | data("read_pos20_6546") |
data("metdt_pos20_6546") |
| Positive | 40 | data("read_pos40_6546") |
data("metdt_pos40_6546") |
| Positive | 60 | data("read_pos60_6546") |
data("metdt_pos60_6546") |
| Positive | 80 | data("read_pos80_6546") |
data("metdt_pos80_6546") |
| Negative | 20 | data("read_neg20_6546") |
data("metdt_neg20_6546") |
| Negative | 40 | data("read_neg40_6546") |
data("metdt_neg40_6546") |
| Negative | 60 | data("read_neg60_6546") |
data("metdt_neg60_6546") |
| Negative | 80 | data("read_neg80_6546") |
data("metdt_neg80_6546") |
PhenolicsDB metadata for GNPS .mgf format
You can also use the metdt_GNPS_pos and
metdt_GNPS_pos data frames to export this library in a
.mgf format.
List of metabolites
In the extdata directory you will find all the
.mzML files for all the standards, as well as the structure
tables to be used jointly with MS2extract.
In the following table you can see the list of metabolites we are including in this version of the database, as well as in what batch you will find them.
Disclaimer: Metabolites with no reported data file have not MS/MS data
| Name | Formula | Monoisotopic_mass | rt (s) |
|---|---|---|---|
| 3-Hydroxybenzaldehyde | C7H6O2 | 122.0368 | 159 |
| 4-Hydroxybenzaldehyde | C7H6O2 | 122.0368 | 154 |
| Malic acid | C4H6O5 | 134.0215 | 29 |
| Salicylic acid | C7H6O3 | 138.0317 | 190 |
| D-(-)-Citramalic acid | C5H8O5 | 148.0372 | 52 |
| (E)-Cinnamic acid | C9H8O2 | 148.0524 | 221 |
| 2,4,6-Trihydroxybenzaldehyde | C7H6O4 | 154.0266 | 158 |
| 2-3-Dihydroxybenzoic acid | C7H6O4 | 154.0266 | 140 |
| 2-5-Dihydroxybenzoic acid | C7H6O4 | 154.0266 | 130 |
| 2-6-Dihydroxybenzoic acid | C7H6O4 | 154.0266 | 135 |
| 3-4-Dihydroxybenzoic acid | C7H6O4 | 154.0266 | 105 |
| Vanillyl alcohol | C8H10O3 | 154.0630 | 120 |
| p-Coumaric acid | C9H8O3 | 164.0473 | 163 |
| Vanillic acid | C8H8O4 | 168.0423 | 140 |
| Gallic acid | C7H6O5 | 170.0215 | 70 |
| Ascorbic acid | C6H8O6 | 176.0321 | 20 |
| Esculetin | C9H6O4 | 178.0266 | 142 |
| Caffeic acid | C9H8O4 | 180.0423 | 141 |
| Glucose | C6H12O6 | 180.0634 | 21 |
| D-Sorbitol | C6H14O6 | 182.0790 | 21 |
| Isoscopoletin | C10H8O4 | 192.0423 | 168 |
| Scopoletin | C10H8O4 | 192.0423 | 173 |
| Quinic acid | C7H12O6 | 192.0634 | 22 |
| Ferrulic acid | C10H10O4 | 194.0579 | 172 |
| Sinapic acid | C11H12O5 | 224.0685 | 174 |
| Apigenin | C15H10O5 | 270.0528 | 236 |
| Naringenin | C15H12O5 | 272.0685 | 183 |
| Phloretin | C15H14O5 | 274.0841 | 236 |
| Fisetin | C15H10O6 | 286.0477 | 197 |
| Kaempferol | C15H10O6 | 286.0477 | 242 |
| (-)-Epicatechin | C15H14O6 | 290.0790 | 142 |
| Catechin | C15H14O6 | 290.0790 | 132 |
| Kaempferide | C16H12O6 | 300.0634 | 293 |
| Ellagic acid | C14H6O8 | 302.0063 | 172 |
| Herbacetin | C15H10O7 | 302.0427 | 212 |
| Quercetin | C15H10O7 | 302.0427 | 220 |
| 3-O-methyl Quercetin | C16H12O7 | 316.0583 | 224 |
| Isorhamnetin | C16H12O7 | 316.0583 | 246 |
| Tamarixetin | C16H12O7 | 316.0583 | 245 |
| Sucrose | C12H22O11 | 342.1162 | 25 |
| Chlorogenic acid | C16H18O9 | 354.0951 | 135 |
| Cryptochlorogenic acid | C16H18O9 | 354.0951 | 136 |
| Neochlorogenic acid | C16H18O9 | 354.0951 | 120 |
| Avicularin | C20H18O11 | 434.0849 | 178 |
| Guaijaverin | C20H18O11 | 434.0849 | 179 |
| Phloridzin | C21H24O10 | 436.1369 | 191 |
| Trilobatin | C21H24O10 | 436.1369 | 201 |
| Astragalin | C21H20O11 | 448.1006 | 182 |
| Quercitrin | C21H20O11 | 448.1006 | 182 |
| Cyanidin 3-Galactoside | C21H21O11 | 449.1084 | 128 |
| Eriodictyol 7-O-glucoside | C21H22O11 | 450.1162 | 169 |
| Ursolic acid | C30H48O3 | 456.3603 | 434 |
| Isoquercitrin | C21H20O12 | 464.0955 | 174 |
| Isoquercitroside | C21H20O12 | 464.0955 | 170 |
| Quercetin-3-galactoside | C21H20O12 | 464.0955 | 174 |
| Quercetin-3-glucoside | C21H20O12 | 464.0955 | 174 |
| Corosolic acid | C30H48O4 | 472.3553 | 371 |
| Quercetin-3-O-glucuronide | C21H18O13 | 478.0747 | 172 |
| Melezitose | C18H32O16 | 504.1690 | 24 |
| Procyanidin A2 | C30H24O12 | 576.1268 | 172 |
| Procyanidin B1 | C30H26O12 | 578.1424 | 118 |
| Procyanidin B2 | C30H26O12 | 578.1424 | 139 |
| Procyanidin B3 | C30H26O12 | 578.1424 | 124 |
| Naringin | C27H32O14 | 580.1792 | 182 |
| Procyanidin | C30H26O13 | 594.1373 | 160 |
| Nictoflorin | C27H30O15 | 594.1585 | 178 |
| Saponarin | C27H30O15 | 594.1585 | 151 |
| Eriocitrin | C27H32O15 | 596.1741 | 166 |
| Neoeriocitrin | C27H32O15 | 596.1741 | 168 |
| Rutin | C27H30O16 | 610.1534 | 166 |
| Narcissin | C28H32O16 | 624.1690 | 180 |
| Quercetin 3-O-sophoroside | C27H30O17 | 626.1483 | 154 |
| Procyanidin C1 | C45H38O18 | 866.2058 | 146 |
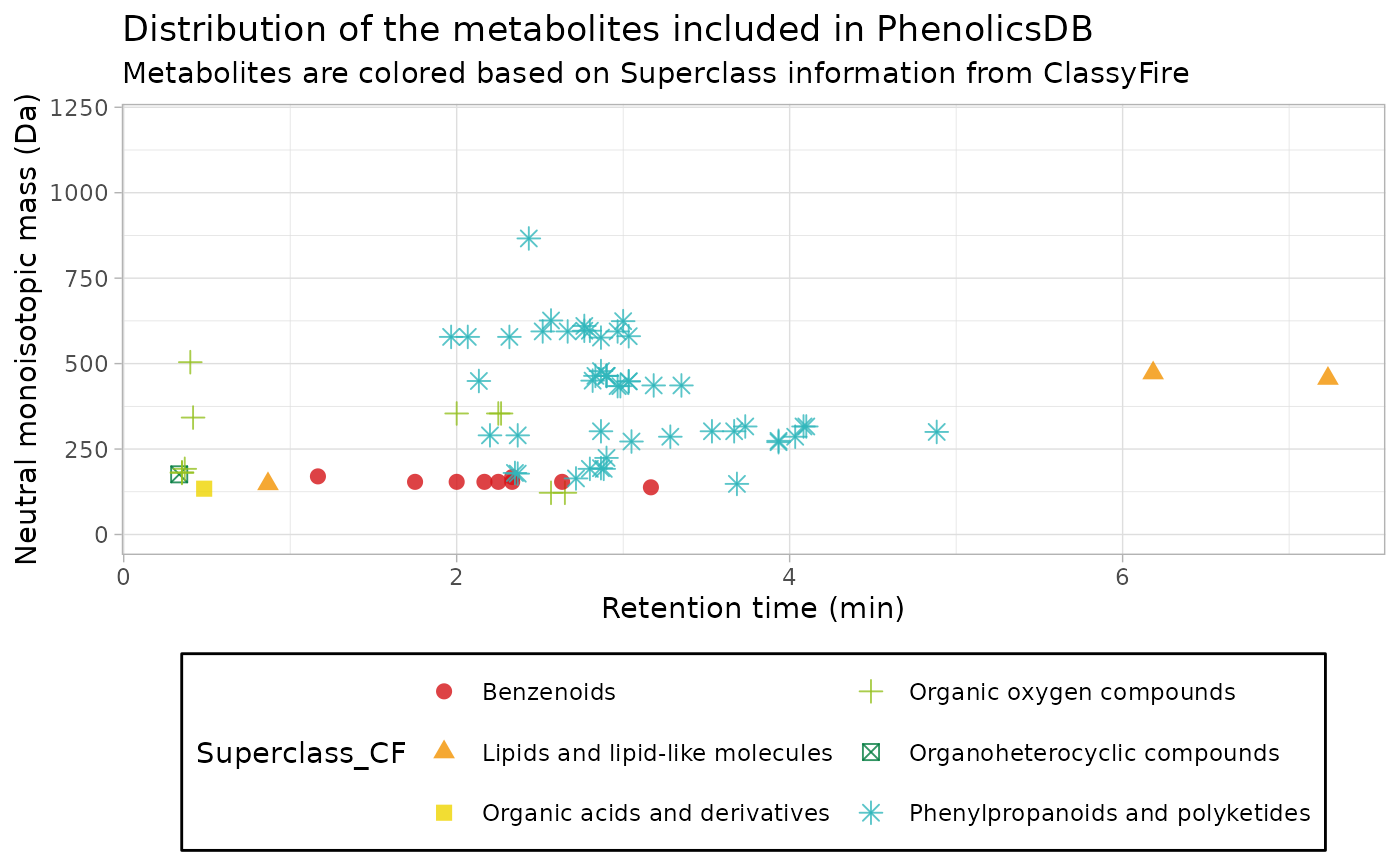
Metabolites in two dimensions
As it was explained above, the chromatography method employed for the LC separation is described in Bilbrey at al. (2022).
Therefore, we can display where each metabolite elutes in terms of retention time and m/z.
Information about this vignette
Code for creating the vignette
## Create the vignette
library("rmarkdown")
system.time(render("Content.Rmd", "BiocStyle::html_document"))
## Extract the R code
library("knitr")
knit("Content.Rmd", tangle = TRUE)Date the vignette was generated.
#> [1] "2025-08-25 19:52:04 UTC"Wallclock time spent generating the vignette.
#> Time difference of 1.681 secsR session information.
#> ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.5.1 (2025-06-13)
#> os Ubuntu 24.04.2 LTS
#> system x86_64, linux-gnu
#> ui X11
#> language en
#> collate C.UTF-8
#> ctype C.UTF-8
#> tz UTC
#> date 2025-08-25
#> pandoc 3.1.11 @ /opt/hostedtoolcache/pandoc/3.1.11/x64/ (via rmarkdown)
#> quarto NA
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> backports 1.5.0 2024-05-23 [1] RSPM
#> bibtex 0.5.1 2023-01-26 [1] RSPM
#> BiocManager 1.30.26 2025-06-05 [1] RSPM
#> BiocStyle * 2.36.0 2025-04-15 [1] Bioconduc~
#> bookdown 0.44 2025-08-21 [1] RSPM
#> bslib 0.9.0 2025-01-30 [1] RSPM
#> cachem 1.1.0 2024-05-16 [1] RSPM
#> cli 3.6.5 2025-04-23 [1] RSPM
#> desc 1.4.3 2023-12-10 [1] RSPM
#> digest 0.6.37 2024-08-19 [1] RSPM
#> dplyr * 1.1.4 2023-11-17 [1] RSPM
#> evaluate 1.0.4 2025-06-18 [1] RSPM
#> farver 2.1.2 2024-05-13 [1] RSPM
#> fastmap 1.2.0 2024-05-15 [1] RSPM
#> fs 1.6.6 2025-04-12 [1] RSPM
#> generics 0.1.4 2025-05-09 [1] RSPM
#> ggplot2 * 3.5.2 2025-04-09 [1] RSPM
#> ggsci 3.2.0 2024-06-18 [1] RSPM
#> glue 1.8.0 2024-09-30 [1] RSPM
#> gtable 0.3.6 2024-10-25 [1] RSPM
#> htmltools 0.5.8.1 2024-04-04 [1] RSPM
#> httr 1.4.7 2023-08-15 [1] RSPM
#> jquerylib 0.1.4 2021-04-26 [1] RSPM
#> jsonlite 2.0.0 2025-03-27 [1] RSPM
#> knitr 1.50 2025-03-16 [1] RSPM
#> labeling 0.4.3 2023-08-29 [1] RSPM
#> lifecycle 1.0.4 2023-11-07 [1] RSPM
#> lubridate 1.9.4 2024-12-08 [1] RSPM
#> magrittr 2.0.3 2022-03-30 [1] RSPM
#> PhenolicsDB * 0.04.3 2025-08-25 [1] local
#> pillar 1.11.0 2025-07-04 [1] RSPM
#> pkgconfig 2.0.3 2019-09-22 [1] RSPM
#> pkgdown 2.1.3 2025-05-25 [1] any (@2.1.3)
#> plyr 1.8.9 2023-10-02 [1] RSPM
#> purrr 1.1.0 2025-07-10 [1] RSPM
#> R6 2.6.1 2025-02-15 [1] RSPM
#> ragg 1.4.0 2025-04-10 [1] RSPM
#> RColorBrewer 1.1-3 2022-04-03 [1] RSPM
#> Rcpp * 1.1.0 2025-07-02 [1] RSPM
#> Rdisop * 1.68.0 2025-04-15 [1] Bioconduc~
#> RefManageR * 1.4.0 2022-09-30 [1] RSPM
#> rlang 1.1.6 2025-04-11 [1] RSPM
#> rmarkdown 2.29 2024-11-04 [1] RSPM
#> sass 0.4.10 2025-04-11 [1] RSPM
#> scales 1.4.0 2025-04-24 [1] RSPM
#> sessioninfo * 1.2.3 2025-02-05 [1] RSPM
#> stringi 1.8.7 2025-03-27 [1] RSPM
#> stringr 1.5.1 2023-11-14 [1] RSPM
#> systemfonts 1.2.3 2025-04-30 [1] RSPM
#> textshaping 1.0.1 2025-05-01 [1] RSPM
#> tibble 3.3.0 2025-06-08 [1] RSPM
#> tidyr 1.3.1 2024-01-24 [1] RSPM
#> tidyselect 1.2.1 2024-03-11 [1] RSPM
#> timechange 0.3.0 2024-01-18 [1] RSPM
#> vctrs 0.6.5 2023-12-01 [1] RSPM
#> withr 3.0.2 2024-10-28 [1] RSPM
#> xfun 0.53 2025-08-19 [1] RSPM
#> xml2 1.4.0 2025-08-20 [1] RSPM
#> yaml 2.3.10 2024-07-26 [1] RSPM
#>
#> [1] /home/runner/work/_temp/Library
#> [2] /opt/R/4.5.1/lib/R/site-library
#> [3] /opt/R/4.5.1/lib/R/library
#> * ── Packages attached to the search path.
#>
#> ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────Bibliography
This vignette was generated using BiocStyle (Oleś, 2025) with knitr (Xie, 2025) and rmarkdown (Allaire, Xie, Dervieux, McPherson, Luraschi, Ushey, Atkins, Wickham, Cheng, Chang, and Iannone, 2024) running behind the scenes.
Citations made with RefManageR (McLean, 2017).
[1] J. Allaire, Y. Xie, C. Dervieux, et al. rmarkdown: Dynamic Documents for R. R package version 2.29. 2024. URL: https://github.com/rstudio/rmarkdown.
[2] M. W. McLean. “RefManageR: Import and Manage BibTeX and BibLaTeX References in R”. In: The Journal of Open Source Software (2017). DOI: 10.21105/joss.00338.
[3] A. Oleś. BiocStyle: Standard styles for vignettes and other Bioconductor documents. R package version 2.36.0. 2025. DOI: 10.18129/B9.bioc.BiocStyle. URL: https://bioconductor.org/packages/BiocStyle.
[4] Y. Xie. knitr: A General-Purpose Package for Dynamic Report Generation in R. R package version 1.50. 2025. URL: https://yihui.org/knitr/.