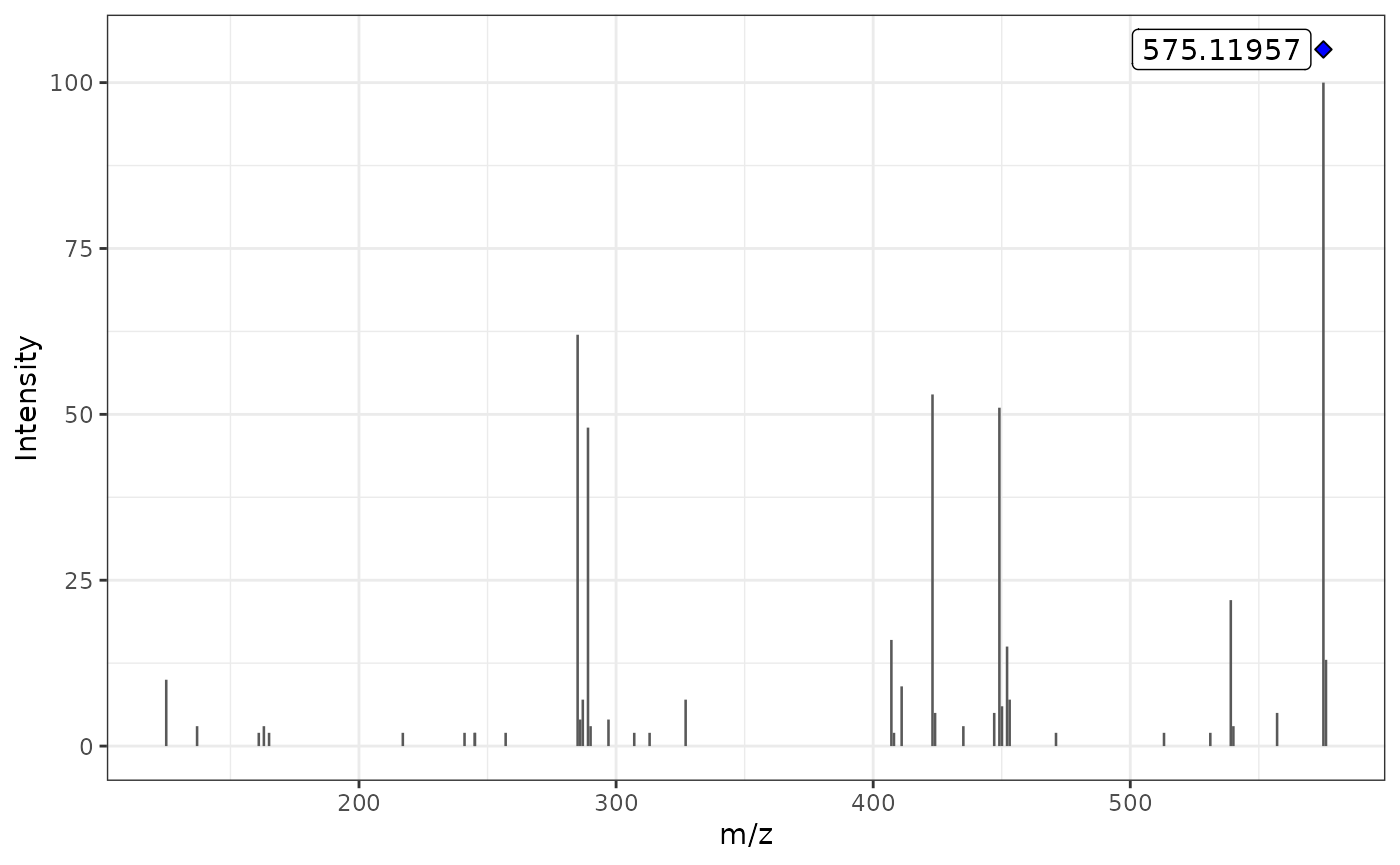
2 - Using the MS2extract batch pipeline
Cristian Quiroz-Moreno & Jessica Cooperstone
2023-08-26
Source:vignettes/using_batch_extract.Rmd
using_batch_extract.RmdIntroduction
In the previous tutorial Introduction to MS2extract package, we described in a detailed manner the core functions of the package. If you are starting to use the MS2extract package with this tutorial, we encourage you to take a look at this tutorial first.
Once you are familiar with the core workflow and functions of this
package, we can dive into an automated pipeline with the proposed
batch_*() functions. If you find that you want to extract
many MS/MS spectra at once, you will want to use
thesebatch_*() functions
The first three main steps have a separate batch_*()
alternative functions; importing mzXML files, extracting MS/MS spectra,
and detecting masses. However, exporting your library to a .msp file is
able to detect if the provided spectra comes from a single or multiple
.mzXML file, so the same function works in both cases.
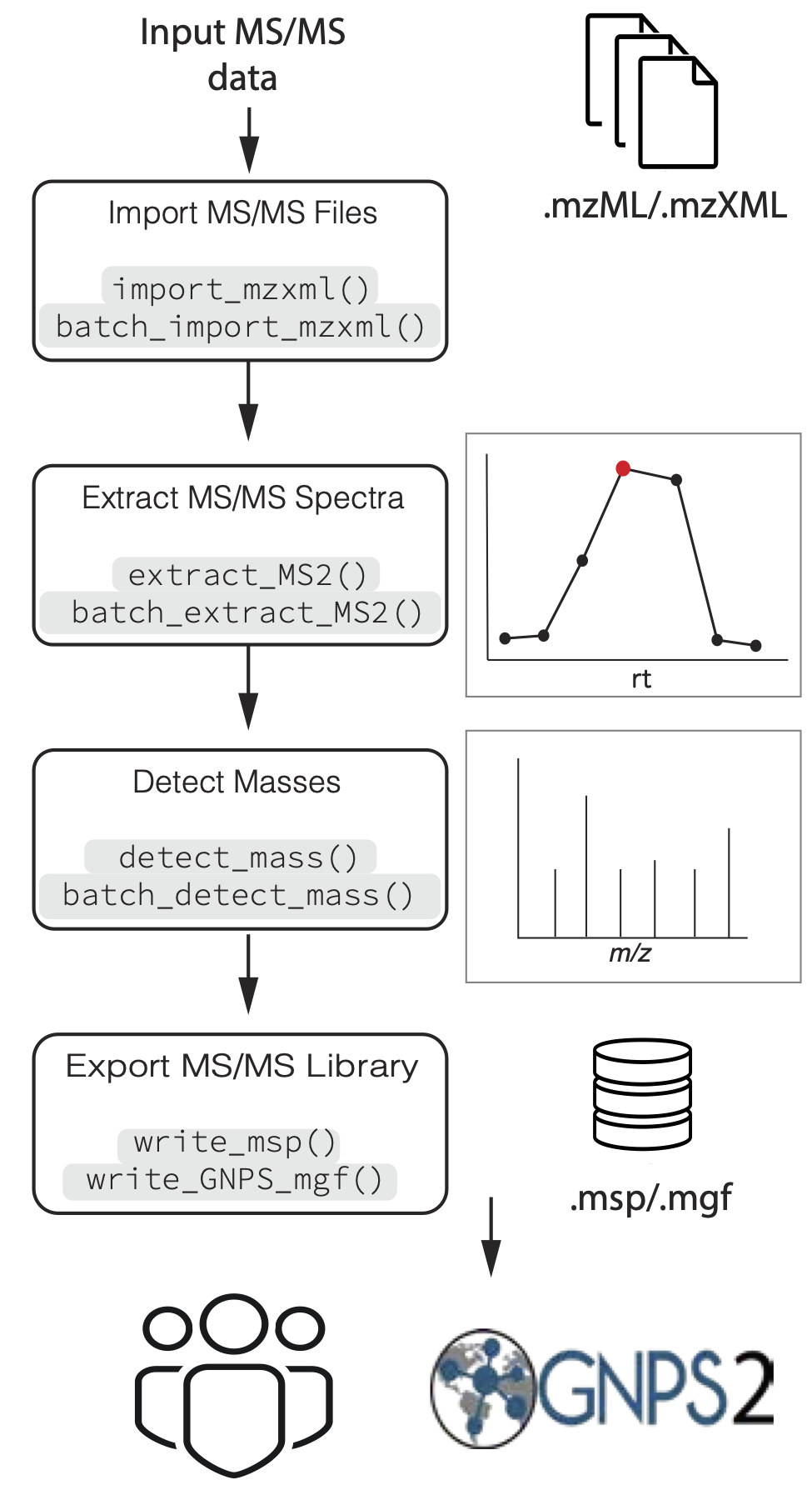
Figure 1. Overview of general data processing pipeline to extract MS/MS spectra using the MS2extract package
Batch functions
We are familiar with the arguments that the core functions accept,
here in this section we describe extra arguments that specific
batch_*() functions require.
batch_import_mzxml
knitr::opts_chunk$set(warning = FALSE)
library(MS2extract)Similarly to import_mzxml(), we need to provide compound
metadata, with at minimum the compound name, formula, ionization mode,
and optionally (but recommended) the region of interest (min_rt
and max_rt).
# Select the csv file name and path
batch_file <- system.file("extdata", "batch_read.csv",
package = "MS2extract"
)
# Read the data frame
batch_data <- read.csv(batch_file)
# File paths for Procyanidin A2 and Rutin
ProcA2_file <- system.file("extdata",
"ProcyanidinA2_neg_20eV.mzXML",
package = "MS2extract"
)
Rutin_file <- system.file("extdata",
"Rutin_neg_20eV.mzXML",
package = "MS2extract"
)
# Add file path - User should specified the file path -
batch_data$File <- c(ProcA2_file, Rutin_file)
# Checking batch_data data frame
dplyr::glimpse(batch_data)
#> Rows: 2
#> Columns: 6
#> $ Name <chr> "Procyanidin A2", "Rutin"
#> $ Formula <chr> "C30H24O12", "C27H30O16"
#> $ Ionization_mode <chr> "Negative", "Negative"
#> $ min_rt <int> 163, 162
#> $ max_rt <int> 180, 171
#> $ File <chr> "/home/runner/work/_temp/Library/MS2extract/extdata/Pr…The only difference between batch_import_mzxml() and
import_mzxml() is that met_metadata can be more than one
row. Here we are working with two compounds, procyanidin A2 and
rutin.
Tip: you can extract multiple compounds from the same .mzXML if they have different precursor ion m/z.
Tip: you can also specify multiple compounds that have the same m/z as long as they have different retention time.
batch_compounds <- batch_import_mzxml(batch_data)
#> Reading MS2 data from ProcyanidinA2_neg_20eV.mzXML
#> Processing...
#> Reading MS2 data from Rutin_neg_20eV.mzXML
#> Processing...The raw mzXML data contains:
- Procyanidin A2: 24249 ions
- Rutin: 22096 ions
# Checking dimension by compound
purrr::map(batch_compounds, dim)
#> $`Procyanidin A2`
#> [1] 24249 4
#>
#> $Rutin
#> [1] 22096 4batch_extract_MS2
Now that we have our data in imported, we can proceed to extract the
most intense MS/MS scan for each compound. In this case, the
batch_extract_MS2() functions do not have extra arguments,
although most of the arguments remains fairly similar.
# Use extract batch extract_MS2
batch_extracted <- batch_extract_MS2(batch_compounds,
verbose = TRUE,
out_list = FALSE
)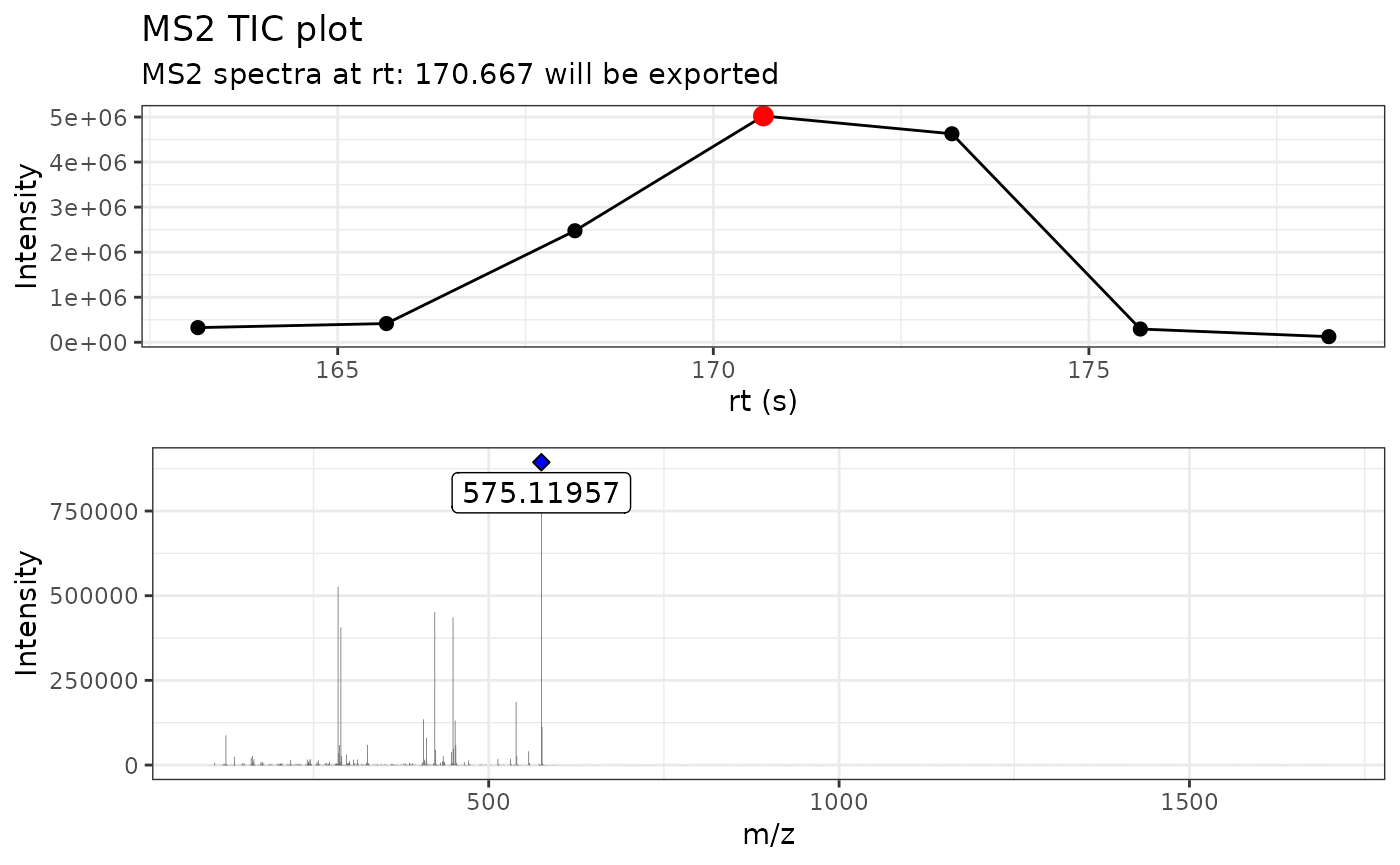
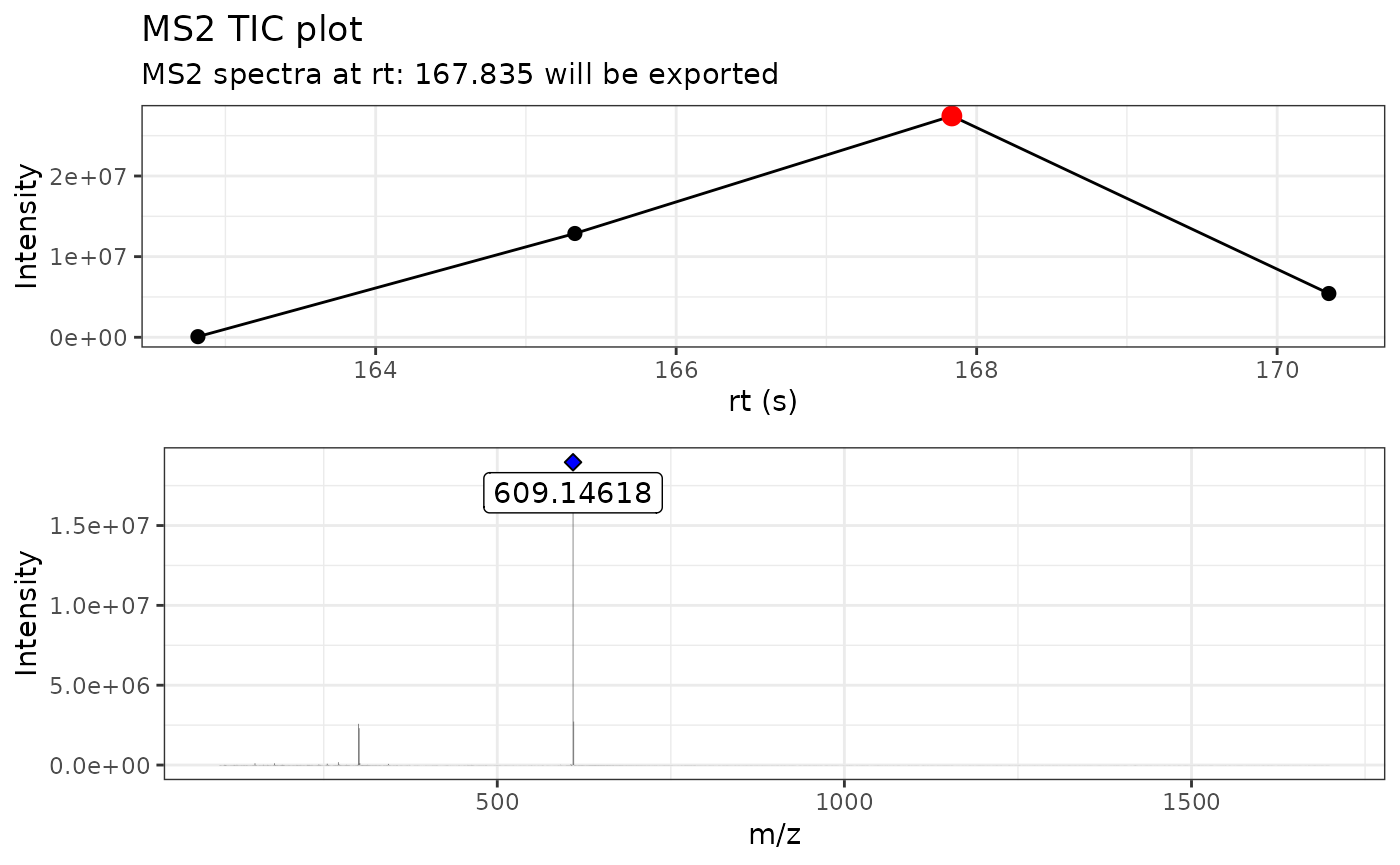
By using verbose = TRUE, we can display the MS/MS TIC
plot as well the raw MS/MS spectra.
batch_detect_mass
Now that we have the raw MS/MS spectra, we are going to remove
background noise/non-informative product ions based on intensity.
batch_detect_mass() has the same arguments than its core
analogue.
batch_mass_detected <- batch_detect_mass(batch_extracted, # Compound list
normalize = TRUE, # Normalize
min_int = 1
) # Minimum intensity
purrr::map(batch_mass_detected, dim)
#> $`Procyanidin A2`
#> [1] 38 4
#>
#> $Rutin
#> [1] 4 4We see a decrease of number of ions, 38 and 4 ions for procyanidin A2 and rutin, respectively.
write_msp
In contrast with the previous batch functions,
write_msp() is able to detect if the user is providing a
single spectra or multiple spectra. However, the user needs to provide
metadata about each compound to be included in the resulting .msp
database.
# Reading batch metadata
metadata_msp_file <- system.file("extdata",
"batch_msp_metadata.csv",
package = "MS2extract"
)
metadata_msp <- read.csv(metadata_msp_file)
dplyr::glimpse(metadata_msp)
#> Rows: 2
#> Columns: 8
#> $ NAME <chr> "Procyanidin A2", "Rutin"
#> $ PRECURSORTYPE <chr> "[M-H]-", "[M-H]-"
#> $ FORMULA <chr> "C30H24O12", "C27H30O16"
#> $ INCHIKEY <chr> "NSEWTSAADLNHNH-LSBOWGMISA-N", "IKGXIBQEEMLURG-NVPNHPE…
#> $ SMILES <chr> "C1C(C(OC2=C1C(=CC3=C2C4C(C(O3)(OC5=CC(=CC(=C45)O)O)C6…
#> $ IONMODE <chr> "Negative", "Negative"
#> $ INSTRUMENTTYPE <chr> "LC-ESI-QTOF", "LC-ESI-QTOF"
#> $ COLLISIONENERGY <chr> "20 eV", "20 eV"After having the cleaned MS/MS spectra and the compound metadata, we can proceed to export them into a .msp file.
write_msp(
spec = batch_mass_detected,
spec_metadata = metadata_msp,
msp_name = "ProcA2_Rutin_batch.msp"
)Session info
sessionInfo()
#> R version 4.3.1 (2023-06-16)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 22.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] MS2extract_0.01.0
#>
#> loaded via a namespace (and not attached):
#> [1] tidyselect_1.2.0 farver_2.1.1 dplyr_1.1.2
#> [4] fastmap_1.1.1 XML_3.99-0.14 digest_0.6.33
#> [7] lifecycle_1.0.3 cluster_2.1.4 ProtGenerics_1.32.0
#> [10] magrittr_2.0.3 compiler_4.3.1 rlang_1.1.1
#> [13] sass_0.4.7 tools_4.3.1 utf8_1.2.3
#> [16] yaml_2.3.7 knitr_1.43 ggsignif_0.6.4
#> [19] labeling_0.4.2 plyr_1.8.8 abind_1.4-5
#> [22] BiocParallel_1.34.2 withr_2.5.0 purrr_1.0.2
#> [25] BiocGenerics_0.46.0 desc_1.4.2 grid_4.3.1
#> [28] stats4_4.3.1 preprocessCore_1.62.1 fansi_1.0.4
#> [31] ggpubr_0.6.0 colorspace_2.1-0 ggplot2_3.4.3
#> [34] scales_1.2.1 iterators_1.0.14 MASS_7.3-60
#> [37] cli_3.6.1 crayon_1.5.2 mzR_2.34.1
#> [40] rmarkdown_2.24 ragg_1.2.5 generics_0.1.3
#> [43] Rdisop_1.60.0 ncdf4_1.21 cachem_1.0.8
#> [46] affy_1.78.2 stringr_1.5.0 zlibbioc_1.46.0
#> [49] parallel_4.3.1 impute_1.74.1 BiocManager_1.30.22
#> [52] vsn_3.68.0 vctrs_0.6.3 carData_3.0-5
#> [55] jsonlite_1.8.7 car_3.1-2 IRanges_2.34.1
#> [58] S4Vectors_0.38.1 ggrepel_0.9.3 MALDIquant_1.22.1
#> [61] rstatix_0.7.2 clue_0.3-64 systemfonts_1.0.4
#> [64] foreach_1.5.2 limma_3.56.2 tidyr_1.3.0
#> [67] jquerylib_0.1.4 affyio_1.70.0 glue_1.6.2
#> [70] MSnbase_2.26.0 pkgdown_2.0.7 codetools_0.2-19
#> [73] cowplot_1.1.1 stringi_1.7.12 gtable_0.3.4
#> [76] OrgMassSpecR_0.5-3 mzID_1.38.0 munsell_0.5.0
#> [79] tibble_3.2.1 pillar_1.9.0 pcaMethods_1.92.0
#> [82] htmltools_0.5.6 R6_2.5.1 textshaping_0.3.6
#> [85] doParallel_1.0.17 rprojroot_2.0.3 evaluate_0.21
#> [88] lattice_0.21-8 Biobase_2.60.0 highr_0.10
#> [91] backports_1.4.1 memoise_2.0.1 broom_1.0.5
#> [94] bslib_0.5.1 Rcpp_1.0.11 xfun_0.40
#> [97] MsCoreUtils_1.12.0 fs_1.6.3 pkgconfig_2.0.3